Understanding how drugs interact with the human brain remains one of the most significant challenges in neuroscience research and CNS drug development. The blood–brain barrier (BBB), while essential for protecting neural tissue, complicates therapeutic delivery. Meanwhile, the neurovascular unit (NVU)—the interconnected network of neurons, astrocytes, microglia, pericytes, and endothelial cells—drives complex physiological behaviors that traditional in vitro systems rarely replicate.
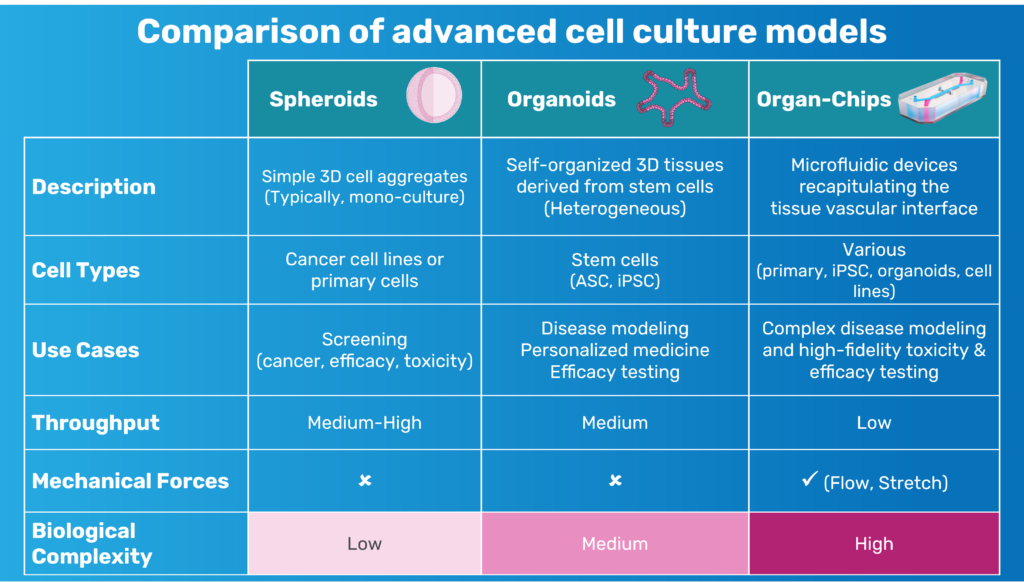
As the field pushes for more predictive, human-relevant models, there has been increasing recognition that the limitations of 2D monolayers, transwell systems, organoids, and animal models constrain our ability to understand BBB transport, neuroinflammation, and human-specific neurovascular function.
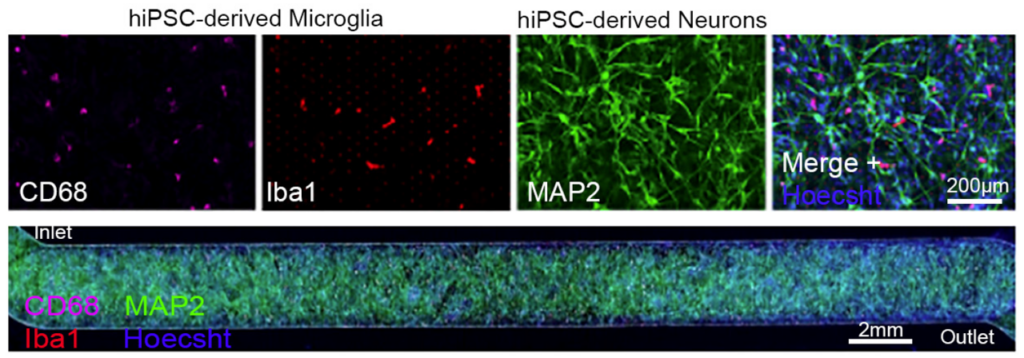
The Emulate Brain-Chip R1 was developed to address these challenges directly. Integrating five iPSC-derived cell types from a single donor within a dynamic Organ-Chip microenvironment, the model captures the cellular diversity, structural organization, and functional interactions that define the human NVU. The result is a system capable of generating more reproducible, mechanistic, and translational insights into human brain physiology.
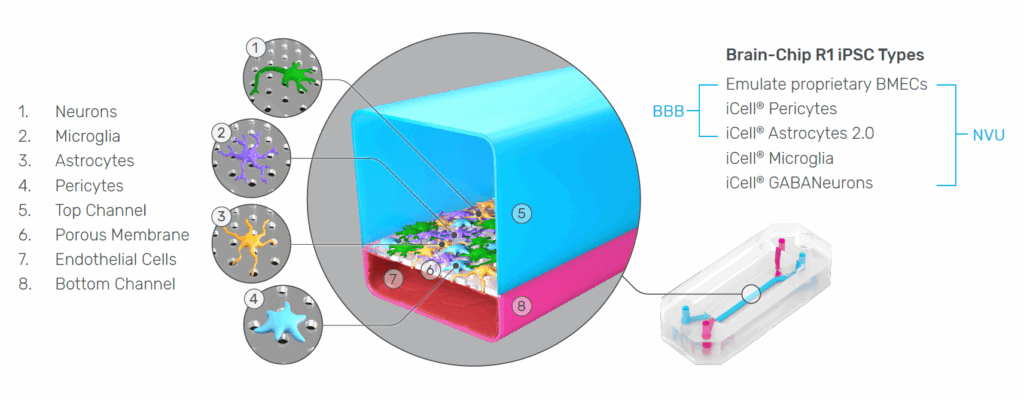
Below, we explore why each of these design features—isogenicity, five-cell composition, and the Organ-Chip foundation—is essential for advancing CNS research, and how the Brain-Chip R1 overcomes longstanding challenges in iPSC model variability and endothelial identity.

1. The Importance of an Isogenic System: Reducing Variability and Increasing Translational Power
A persistent challenge in NVU and BBB modeling is biological variability. When endothelial cells, astrocytes, neurons, microglia, and pericytes come from different donors or differentiation workflows, their genetic and epigenetic differences introduce inconsistencies in phenotype, signaling behavior, and barrier formation. Subtle variations can cascade into meaningful discrepancies in transporter expression, cytokine secretion, and glial activation—especially problematic for studies requiring high precision, such as permeability measurements or efflux transporter assays.
The Brain-Chip R1 eliminates this concern by using five iPSC-derived cell types from a single donor line, forming a truly isogenic system. This allows the NVU’s cellular populations to develop within a shared genetic context, better reflecting the physiological relationships found in vivo.
In practice, isogenicity provides:
- predictable cell–cell communication
- a stable, tight endothelial barrier
- reproducible transporter expression
- a consistent inflammatory baseline
- reduced batch-to-batch variability
For teams advancing CNS therapeutics—where reproducibility and quantification are essential—an isogenic foundation isn’t just an advantage; it’s a requirement.

2. Why All Five NVU Cell Types Matter: Capturing the Biology That Drives BBB and Brain Function
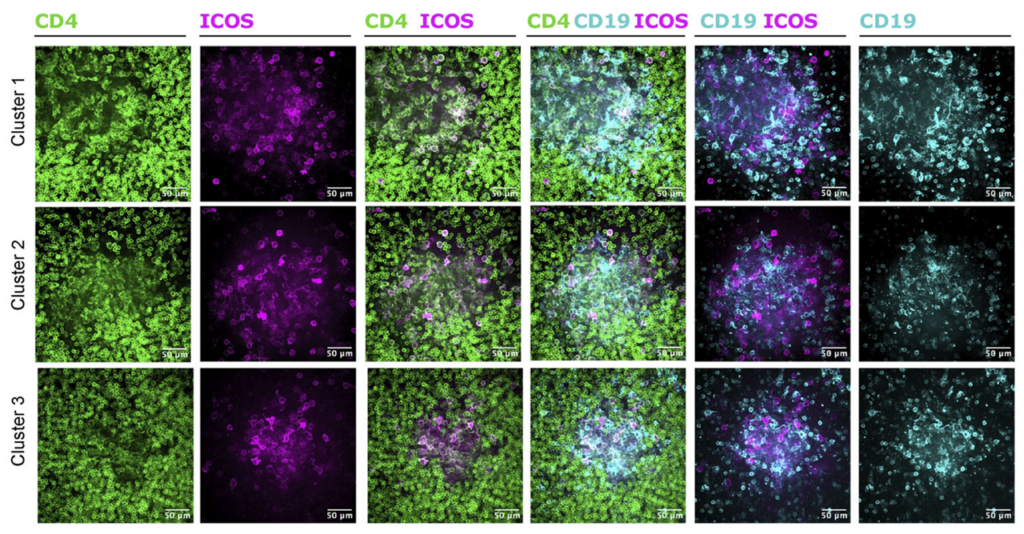
The blood–brain barrier is not a simple endothelial wall—it is the product of coordinated interactions among multiple cell types. In vivo, neurons, astrocytes, microglia, pericytes, and brain microvascular endothelial cells (BMECs) are structurally intertwined and functionally interdependent. Each contributes essential regulatory roles that determine how the BBB forms, how it responds to stimuli, and how substances enter or exit the brain. Recreating this complexity in vitro is therefore fundamental to building a model that behaves like the human NVU.
Endothelial Cells: Gatekeepers of the BBB
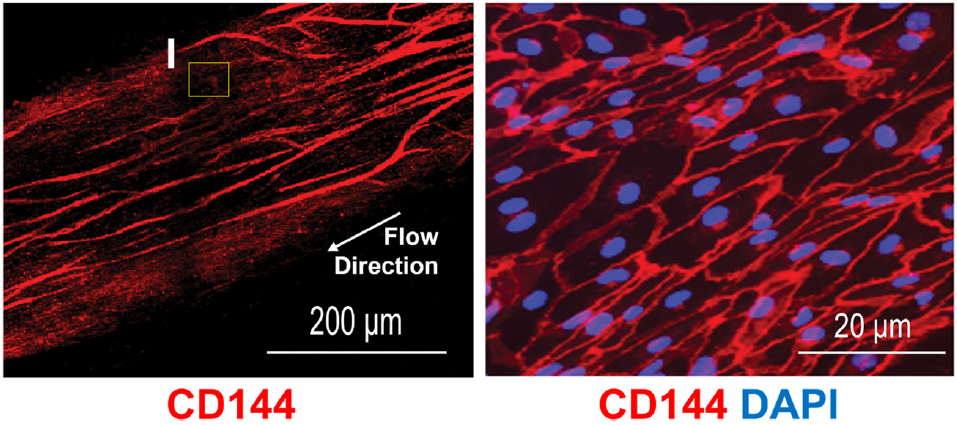
BMECs form the physical barrier that controls molecular entry into the CNS. Their tight junctions, selective permeability, and transporter systems govern nutrient supply, waste clearance, and drug penetration. Without physiologically relevant BMECs, it is impossible to model transport mechanisms that determine whether a therapeutic can reach the brain.
Astrocytes: Inducers and Maintainers of Barrier Function
In vivo, astrocytes regulate BBB tightness, metabolic exchange, and ion homeostasis. Their endfeet envelop capillaries, releasing factors that strengthen endothelial junctions and maintain barrier integrity. Including astrocytes in an in vitro model of the NVU is critical for supporting normal endothelial behavior and for studying barrier breakdown in disease.
Pericytes: Stabilizers of Vascular Structure and Permeability
Pericytes wrap around endothelial cells and play an essential role in vascular stability, angiogenesis, and BBB maturation. They modulate permeability and help maintain the basement membrane. Models lacking pericytes often fail to reproduce realistic barrier tightness or NVU architecture.
Microglia: First Responders of Neuroimmune Signaling
Microglia continuously survey the CNS, shaping synaptic networks and mounting immune responses. Their activation drives cytokine release, inflammation, and barrier disruption during disease. Because neuroinflammation is closely tied to BBB function, incorporating microglia is essential for modeling healthy homeostasis and pathological responses.
Neurons: Essential Targets for CNS Drug Discovery
Neurons are the ultimate therapeutic targets for most CNS drug programs. In vivo, they shape metabolic demand and neurovascular coupling, but in an in vitro NVU model they serve an equally critical purpose: confirming that compounds not only cross the BBB, but also reach and engage their intended targets. Without neurons present, it is impossible to assess whether a drug that successfully penetrates the barrier ultimately affects the cells it is designed to modulate.
Why this matters for BBB and NVU modeling
These five cell types form an ecosystem in the brain where communication—not isolation—drives function. When any component is missing, in vitro models lose essential regulatory pathways: astrocytes cannot tighten the barrier, microglia cannot mount inflammatory responses, neurons cannot signal metabolic demand, and pericytes cannot stabilize vascular integrity.
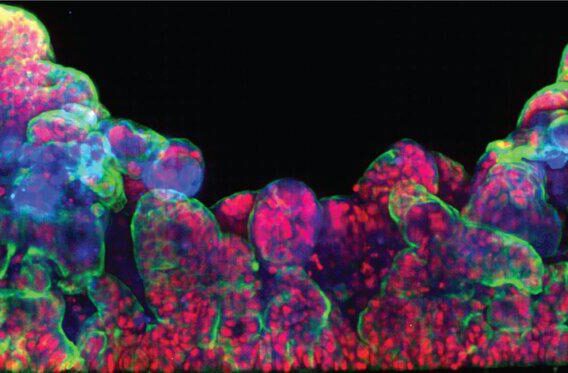
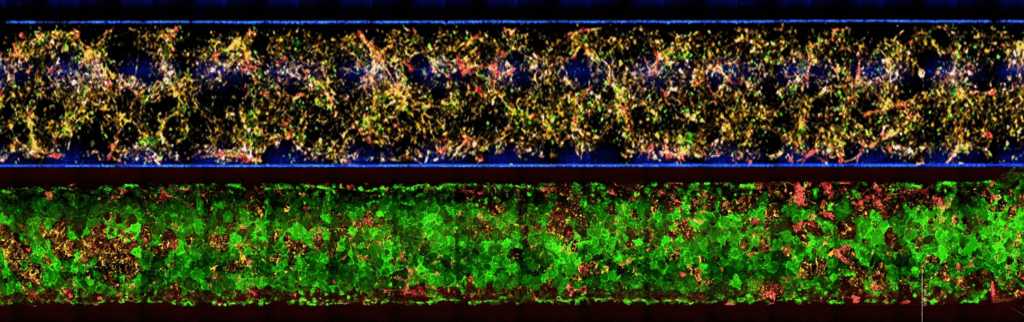
A model containing the full NVU cast offers a more complete and physiologically relevant platform for studying permeability, inflammation, drug transport, and disease mechanisms—ensuring that biological responses arise from the same multicellular interactions that govern the BBB in the human brain. Together, the five cell populations of Brain-Chip R1 organize into integrated multicellular clusters, recreate NVU cytoarchitecture, and support a range of human-relevant functional responses.
Advancing Beyond Conventional iPSC-Derived BMECs
A major limitation of many existing iPSC-derived BBB models is the “epithelialization” problem. Traditional iPSC-based BMEC protocols often generate cells with epithelial-like traits—expressing epithelial markers, forming unnaturally rigid barriers, or lacking key endothelial genes. This misalignment complicates mechanistic transport studies and undermines physiological relevance.
Emulate’s custom BMECs were engineered specifically to overcome these limitations. They express hallmark endothelial markers—including Claudin-5, VE-Cadherin, ZO-1, and PECAM1—while lacking epithelial markers such as EPCAM1 and cytokeratins. Their transporter profile closely mirrors the human BBB, with robust GLUT1, P-gp, and TfR1 expression supporting nutrient transport, efflux activity, and receptor-mediated transcytosis.
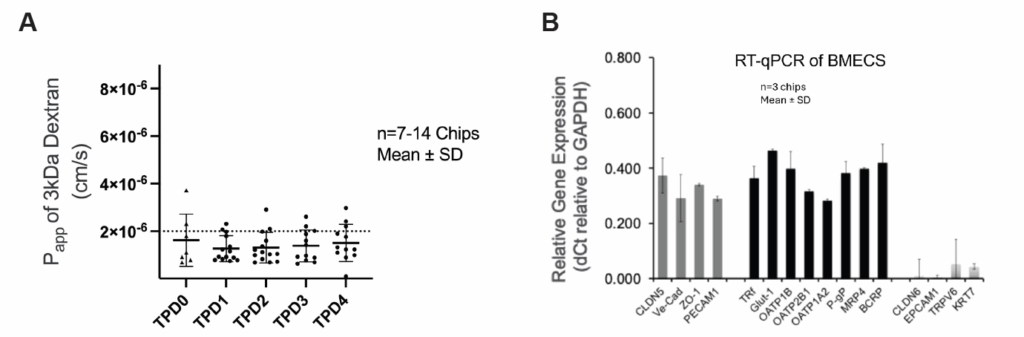
In the Brain-Chip R1, these BMECs form a continuous, tight, and stable monolayer that remains physiologically relevant throughout the experimental window. Their identity and function support quantitative permeability assays and mechanistic transport studies that more accurately reflect in vivo BBB behavior.
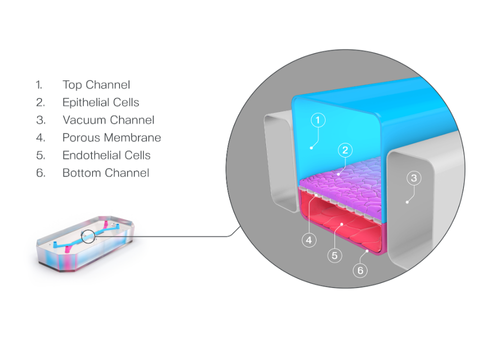
3. The Organ-Chip Platform: Why Microfluidics and Mechanical Cues Matter
Even with the right cell types, the physical environment plays a critical role in determining how the model will behave. Static cultures lack perfusion, shear stress, and compartmentalized architecture—features fundamental to BBB physiology.
The Brain-Chip R1’s dual-channel microfluidic design recreates the capillary–brain interface. The vascular channel experiences controlled media flow, promoting tight junction formation and maintaining proper endothelial polarity. A thin, porous membrane allows astrocyte projections and signaling molecules to reach the endothelium, while compartmentalization mirrors in vivo tissue interfaces.
This dynamic system supports:
- stable barrier formation
- physiologically relevant transporter expression
- nutrient and waste exchange
- cross-compartment signaling
- extended structural integrity
Together, these properties result in a model that much more faithfully recapitulates the in vivo characteristics of the BBB and NVU.

4. Overcoming iPSC Workflow Variability Through a Strategic Partnership of Industry Leaders
iPSC technologies offer tremendous promise, but they are often accompanied by significant variability. Differentiation workflows can be sensitive to subtle changes in reagents or culture conditions, and generating high-quality NVU-relevant cells typically requires specialized expertise. Variability in iPSC-derived astrocytes, microglia, neurons, or pericytes can dramatically alter BBB behavior, leading to inconsistent or difficult-to-reproduce results.
Emulate’s partnership with FUJIFILM Cellular Dynamics, Inc. (FCDI) directly addresses these challenges. FCDI is recognized for its rigorously standardized, industrialized iPSC differentiation processes, which produce stable, high-quality cell populations at scale. By integrating these cells into Emulate’s direct-to-chip workflow, the Brain-Chip R1 delivers the scientific value of iPSC-derived NVU modeling without the operational burden traditionally associated with iPSC platforms.
Researchers gain access to reproducible, well-characterized cell types—eliminating the need to differentiate, quality-control, or scale these populations in-house. Combined with Emulate’s optimized media formulations and microfluidic stability, the partnership ensures a consistent, experiment-ready NVU environment with each run.

5. Enabling Human-Relevant, Mechanistic Insights into Brain Function
The Brain-Chip R1’s combination of isogenic cells, multicellular complexity, and dynamic microfluidics enables a depth of mechanistic insight that traditional in vitro systems cannot match.
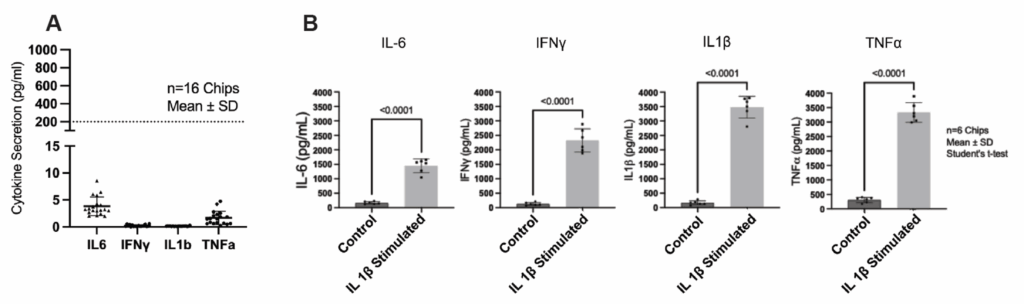
The endothelial barrier maintains consistently low permeability over time, allowing for quantitative evaluation of passive diffusion, carrier-mediated transport, and efflux processes. Meanwhile, glial populations remain in a resting state until stimulated—making it possible to distinguish baseline NVU behavior from induced neuroinflammatory responses.
An additional advantage of the Brain-Chip R1 model is the Chip-R1™ Rigid Chip’s minimally absorbing materials. Where PDMS-based systems can absorb hydrophobic drugs and distort permeability measurements, the Chip-R1 maintains high compound recovery. This makes the Brain-Chip R1 particularly powerful for ADME-relevant assays and quantitative BBB transport studies.
These features support applications including BBB penetrance studies, transport mechanism analysis, ADME assessment, cytokine profiling, neuroinflammation modeling, and disease-related pathway exploration.

Conclusion
The Brain-Chip R1 represents a major step forward for translational neuroscience and CNS drug development. By combining a fully isogenic foundation, five essential NVU cell types, custom BMECs, dynamic microfluidic architecture, low-drug-absorption materials, and standardized workflows, it brings together the most important features required for a human-relevant BBB and NVU model.
Researchers can now explore BBB transport, NVU signaling, neuroinflammation, and brain–vasculature interactions with unprecedented consistency and mechanistic clarity—accelerating discovery and offering a more predictive path toward CNS therapeutic development.
Interested in learning more? Download the Brain-Chip R1 Technical Note to read the see the full data set!