Writing in a recent issue of Nature Reviews Drug Discovery, Jack Scannell, author of Eroom’s law, and a team of experts stated plainly that predictive validity is “the thing that nearly everyone already believes is important [but which] is more important than nearly everyone already believes.” In other words, the drug development industry knows that predictive validity is important, but Scannell and his co-authors argue that it is far more important than most understand it to be.
Predictive validity describes a tool’s ability to reliably predict a future outcome. The drug development industry has struggled for centuries to find preclinical models that accurately predict which drug candidates will be both safe and effective in humans. Despite remarkable advances in science over the past half-century, including the advent of tools like next-generation sequencing and CRISPR editing technology, success in drug development remains rare.
“Successful drug discovery is like finding oases of safety and efficacy in chemical and biological deserts,” writes Scannell and his co-authors, referring to the 90% to 97% clinical trial failure rate that the industry is familiar with. They argue that the drug developers have spent recent decades attempting to improve success rates through brute force scale—the equivalent of searching for oases by simply running over more desert terrain. Instead, they argue, the industry should focus on improving the predictive validity of existing preclinical models, as these are the compasses that will lead to success.
The Faulty Preclinical Models Still In Use
At every stage of the drug discovery and development process, researchers must choose which compounds are worth investing in and which should be left by the wayside. To make these difficult decisions, they rely on tools such as cell lines, animal models, or computer modeling to predict how these prospective compounds will behave in patients. Naturally, the quality of these tools—specifically their predictive validity—has a direct impact on the quality of drug development decisions.
If a model has perfect predictive validity, you can trust that a drug’s behavior in the model will perfectly match its behavior in patients. However, researchers have yet to find such a model—a reality that has greatly limited drug development success.
Take for instance the use of rodents to study ischemic stroke. Such models are easy to use and widely available, making them particularly attractive for drug testing. And they are robust—that is, the performance of compounds in one rodent model is likely to replicate in another.
However, there are important genetic and physiological differences between rodents and humans that severely reduce these models’ predictive validity for drug performance in humans. Not surprisingly, these rodent models select drugs that are both safe and effective for rodents, but not necessarily for humans. Scannell and his co-authors underscore this point by saying that “it now seems likely that non-diabetic normotensive rodents respond better to a range of neuroprotective drugs than do the bulk of elderly human patients with stroke.”
In other words, a rat, no matter how fancy, can only go so far in mimicking human physiology and disease response. In reality, no model can be perfectly predictive. What they can do is provide narrow ranges of value—what Scannell and his co-authors refer to as “domains of validity.”
Stay Up to Date with the Emulate newsletter
Domain of Validity and the Limitations of Models
A domain of validity is the specific context in which a model is most predictive. Take tumor cell lines in oncology, for example. For nearly a century, researchers have been developing cytotoxic drugs to treat cancer based on studies in which human cancer cell lines are grown as 2D monolayers. These cells are typically fast growing, genetically homogenous, and able to be rapidly scaled up for high-throughput screening. However, their fast-growing nature leaves them uniquely susceptible to cytotoxic drugs—something researchers have known since the late 1960s1.
Many cancers, though, comprise heterogeneous and slow-growing cells. It’s no surprise then that between 2000 and 2015, oncology had the highest clinical trial failure rate of any major therapy area at 97%2,3. Human cancer cell lines are valuable tools, but they are representations of the small subset of fast-growing cancers.
Put another way, many cancer cell lines are at their peak predictive validity only in the narrow domain of predicting drug effects in fast-growing, homogenous tumors. By using these cells to represent tumors broadly, researchers unknowingly extended the model well beyond its narrow domain of authority and, as a result, overestimated how effective cytotoxic drugs would be in tumors containing slow-growing cells.
These examples show that it is critical for researchers to carefully consider their models’ predictive limitations when making drug development decisions. They also exemplify that the standard models for preclinical decision-making—namely animals and conventional cell culture—are severely limited, and this has greatly curtailed the pace of drug development and driven the cost of bringing a new drug to market above $2 billion.
Many have tried to compensate for models’ predictive limitations by increasing model throughput, running the same models at a higher pace to gather more data. However, Scannell and his co-authors urge the industry to focus instead on developing better, more predictive models.
Towards Better Model Predictive Validity
In previous papers, Scannell and many of his co-authors have argued that incremental improvements in the predictive validity of a model can have a far greater impact on drug development success than simply improving the number of compounds being screened4,5. Clearly, part of improving predictive validity requires researchers to evaluate their models and establish domains of validity for each.
Unfortunately, doing so is a complex process that often lacks incentives. It may take years or decades to determine if a tool is truly predictive of clinical outcome, and performing such an analysis would require dedicated funds and clear definitions of what qualifies as sufficiently predictive.
Fortunately, Scannell and his co-authors offer several suggestions for how we can improve model validity. An abbreviated list is presented below:
- Evaluate the models we use as best we can. This should include the collection of structured and consistent data on model performance and an explicit effort to both reduce bias and ensure that criteria for evaluation are informed by expert opinions.
- Fund retrospective studies that compare tools to determine why some models are successful and others are not.
- Establish a system for institutional learning such that, after evaluations are performed, others can learn from it and iterate.
- Foster inter-organizational information sharing of detailed protocols, calibration methods, calibration results, decision tools’ contexts-of-use, and associated craft skills.
Improving Drug Development with Organ-Chips
Ultimately, these authors make it clear that the drug development industry—as well as the patients who rely on it—stand to benefit greatly from close examination of models’ predictive validity. Ideally, drug development pipelines would not be static or adherent to models based on tradition, but rather would select the models based on superior and complementary domains of authority.
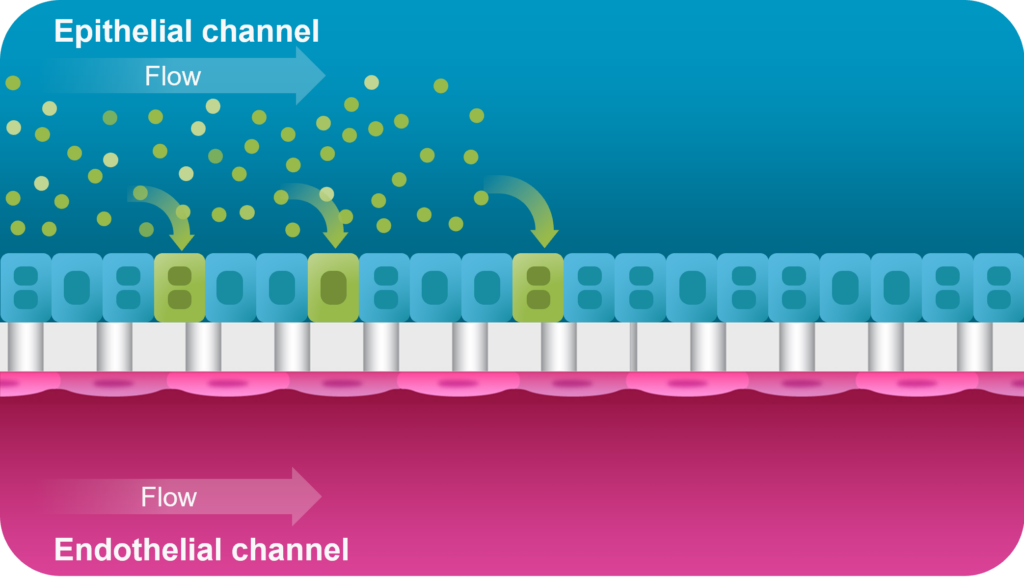
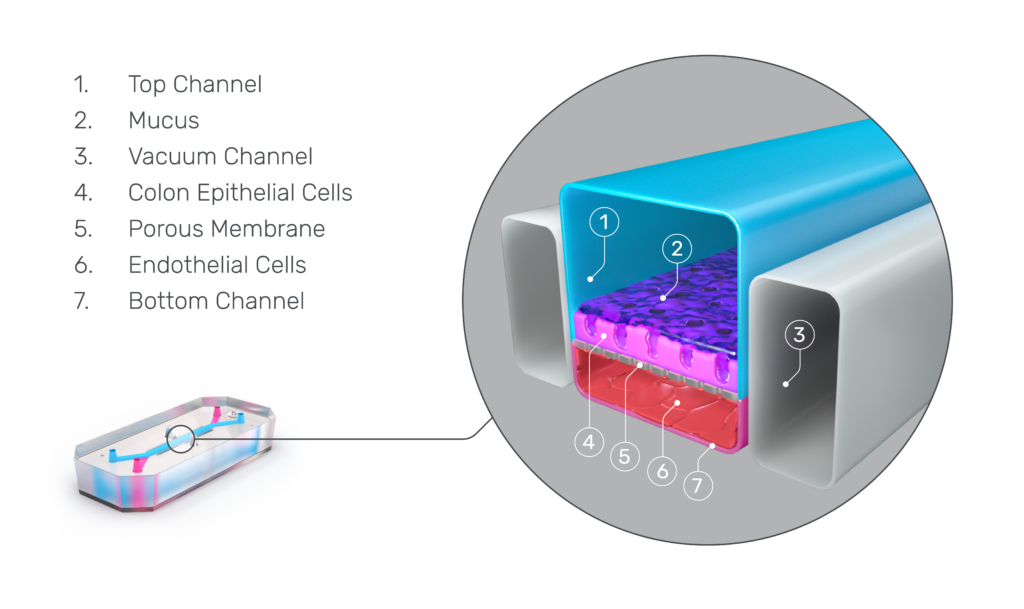
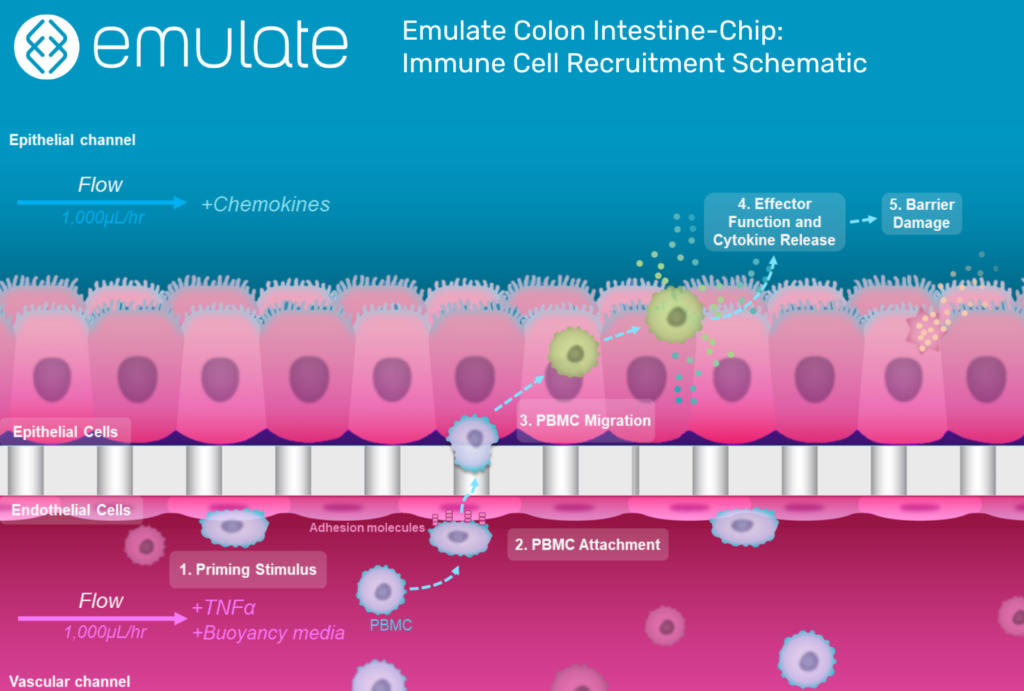
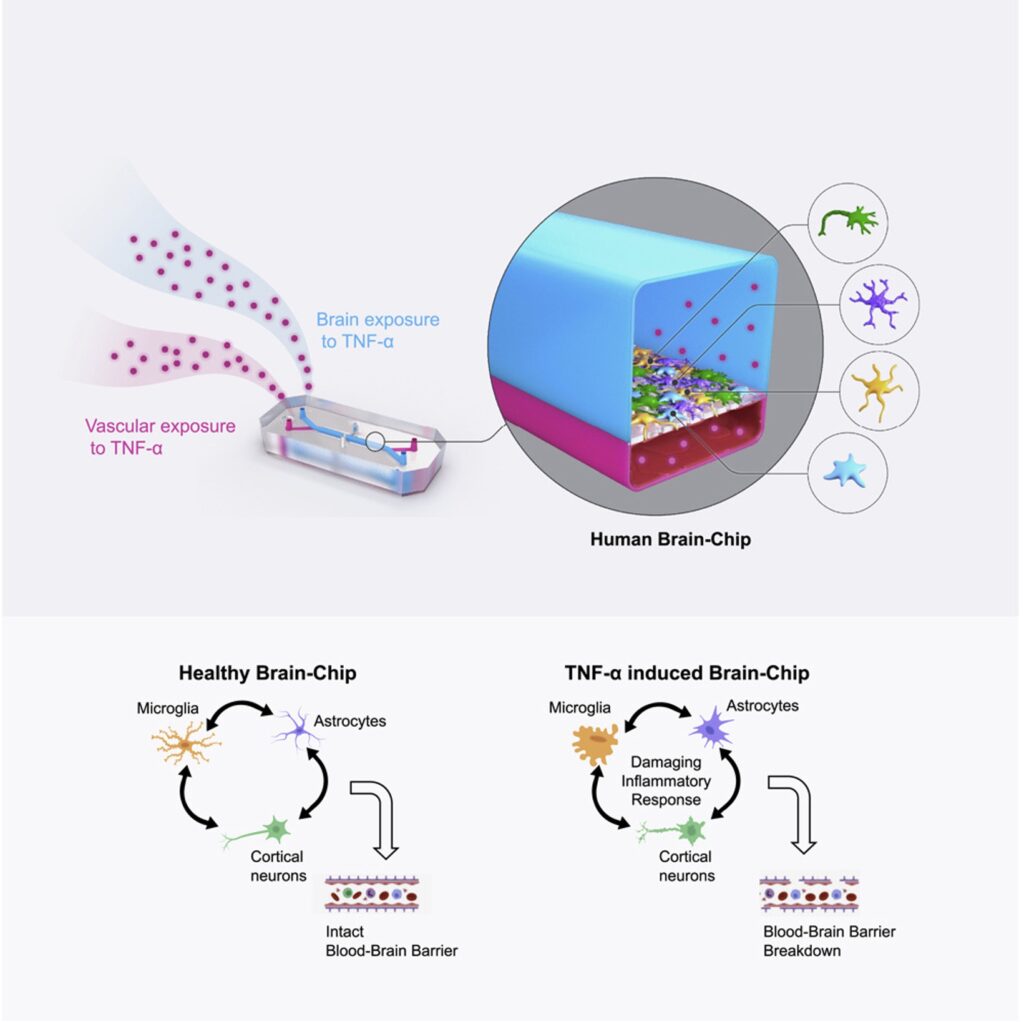
This is where Organ-on-a-Chip technology can make a big impact. Organ-Chips are unique model systems that integrate living human cells, tissue-tissue interactions, tissue-specific proteins, chemical environments, and biomechanical forces to emulate human tissues. Several lines of evidence show that Organ-Chips can closely resemble in vivo human organ behavior and may be beneficial for predicting drug toxicity, among many other applications.
Exemplifying this, a recent paper in Communications Medicine, part of Nature Portfolio, showed that the Emulate human Liver-Chip model was far more predictive of drug-induced liver toxicity relative to animal and spheroid models. In collaboration with Scannell, the team behind the study then showed that integrating the Emulate human Liver-Chip into drug development pipelines could result in more than $3 billion dollars in excess productivity for the drug development industry, owing to the model’s improved predictive validity.
Along with the Emulate human Liver-Chip study, the argument made by Scannell and his co-authors in Nature Reviews Drug Discovery may be a critical moment of reckoning for drug developers. Now is the time to prioritize predictive validity and begin integrating better, more human-relevant tools into drug development programs.
References
- Skipper HE. The effects of chemotherapy on the kinetics of leukemic cell behavior. Cancer Res. 1965 Oct;25(9):1544-50. PMID: 5861078.
- Chabner, B., Roberts, T. Chemotherapy and the war on cancer. Nat Rev Cancer 5, 65–72 (2005). https://doi.org/10.1038/nrc1529
- DiMasi, J.A. (2001), Risks in new drug development: Approval success rates for investigational drugs. Clinical Pharmacology & Therapeutics, 69: 297-307. https://doi.org/10.1067/mcp.2001.115446
- Scannell JW, Bosley J (2016) When Quality Beats Quantity: Decision Theory, Drug Discovery, and the Reproducibility Crisis. PLoS ONE 11(2): e0147215. https://doi.org/10.1371/journal.pone.0147215
- Ringel, M. S., Scannell, J. W., Baedeker, M., & Schulze, U. (2020, April 16). Breaking eroom’s law. Nature News. Retrieved March 21, 2023, from https://www.nature.com/articles/d41573-020-00059-3