Jim Corbett, CEO of Emulate, Inc., released testimony today in support of the FDA Modernization Act of 2021 (H.R 2565 and S. 2952). This submission represents Emulate’s enthusiastic endorsement of the act and recognition that this step is essential for increasing the safety and efficacy of pharmaceutical development. Standards for drug development have remained essentially unchanged since the passage of the Federal Food, Drug, and Cosmetic Act (FFDCA) in 1938, which required all drugs in development to be tested in animals before reaching humans. While this did lead to a safer drug-development process at the time, species differences between animals and humans often mean the results from preclinical research cannot accurately apply to humans. This means that a drug’s toxicity could remain hidden until it reaches human trial or the market.
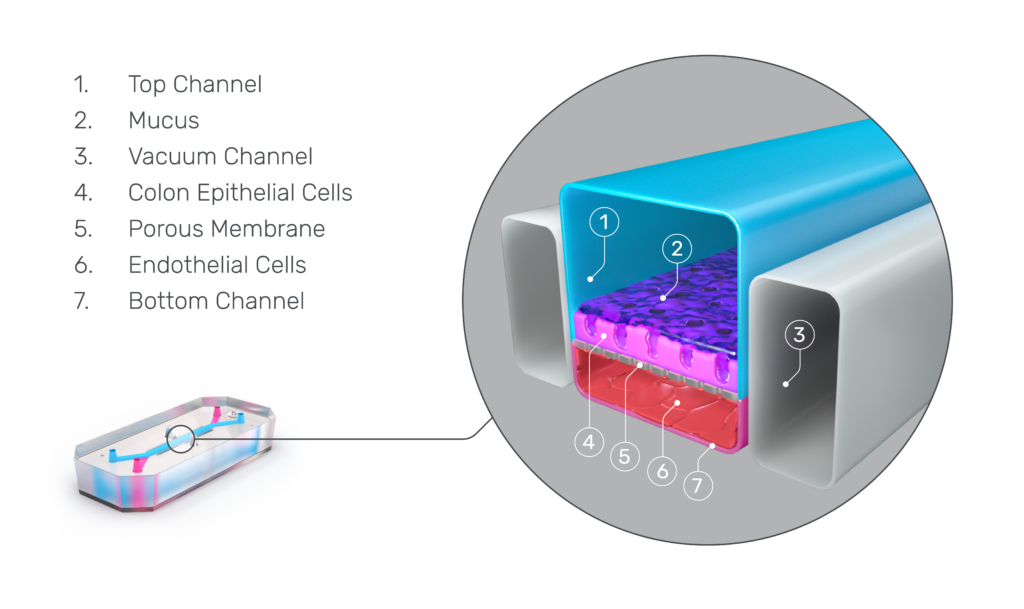
It is for these reasons that Corbett proudly submits testimony in support of the FDA Modernization Act, which will allow sponsor organizations to submit data from studies using microphysiological systems and other non-animal alternatives, such as Emulate’s Organs-on-Chips.
Testimony of
Jim Corbett, CEO Emulate, Inc.
Before the Subcommittee on Health of the Committee on Energy and Commerce Thursday, March 17, 2022
The Future of Medicine: Legislation to Encourage Innovation and Improve Oversight
On behalf of Emulate, Inc., the leading provider of organ-on-a-chip technology, I offer this testimony in support of the FDA Modernization Act of 2021 (H.R 2565 and S. 2952).
Since the Federal Food, Drug, and Cosmetics Act (FFDCA) of 1938 mandated that all new drugs be tested for toxicity in animals prior to human studies, scientific advancements have been abundant. However, predicting human toxicity through animals models still leaves us with a public health issue.
There is no doubt that animal models have contributed to major advances in medicine and have contributed to safe and effective drugs making it to market. However, these models have the difficult job of approximating the human body, and sometimes they get it wrong.
A growing body of evidence suggests that animal models are lacking in both sensitivity and specificity when it comes to predicting drug toxicity in humans.1-3 A 2014 study analyzing the effects of 2,366 drugs in both animals and humans found that “tests on animals (specifically rat, mouse and rabbit models) are highly inconsistent predictors of toxic responses in humans and are little better than what would result merely by chance.”4 A 2008 review found similar results, concluding that animal models predicting drug toxicity in humans may have sensitivity and specificity values below 70%.2
The cost of poor specificity and selectivity is too often passed onto the patient. A review of 578 discontinued and withdrawn drugs in Europe and the United States showed that nearly half halted distribution due to post-approval toxicity.5 Similarly, a 2012 analysis of 43 post-approval drugs with serious toxicity effects found that only 19% of them showed indications of toxicity in animal studies.6
In a recent study published to bioRxiv, researchers found the human Liver-Chip to have an 87% sensitivity and 100% specificity when differentiating hepatotoxic from non-hepatotoxic small molecules.7 Importantly, all 22 hepatotoxic drugs included in the study had previously been classified as safe due to a lack of toxicity in animal models. Collectively, these compounds resulted in 208 patient fatalities and 10 liver transplants. Had the human Liver-Chip been used during preclinical screening of these compounds, it’s likely that many of these fatalities could have been avoided.
Animal models have played an undeniably significant role in the evolution of medicine, and will continue to do so, but to make the drug development process safer, more efficient, and more humane, we must take a hard look at how we can leverage scientific advancements to continuously improve patient safety.
Sincerely,
Jim Corbett
CEO
Emulate, Inc.
- Van Norman GA. Limitations of animal studies for predicting toxicity in clinical trials: Is it time to rethink our current approach? JACC Basic Transl Sci. 2019;4(7):845-854. 2019. doi: 10.1016/j.jacbts.2019.10.008
- Matthews RA. Medical progress depends on animal models – doesn’t it? J R Soc Med. 2008;101(2):95-98. doi: 10.1258/jrsm.2007.070164
- Not-OD-12-025: NIH research involving chimpanzees. National Institutes of Health. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-12-025.html. Accessed December 17, 2021.
- Bailey J, Thew M, Balls M. An analysis of the use of animal models in predicting human toxicology and Drug Safety. Altern Lab Anim. 2014;42(3):181-199. doi: 10.1177/026119291404200306
- Siramshetty VB, Nickel J, Omieczynski C, Gohlke BO, Drwal MN, Preissner R. WITHDRAWN–a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016;44(D1):D1080-D1086. doi: 10.1093/nar/gkv1192
- van Meer PJK, Kooijman M, Gispen-de Wied CC, Moors EHM, Schellekens H. The ability of animal studies to detect serious post marketing adverse events is limited. Regul Toxicol Pharmacol. 2012;64(3):345-349. doi: 10.1016/j.yrtph.2012.09.002
- Ewart, Lorna, et al. Qualifying a Human Liver-Chip for Predictive Toxicology: Performance Assessment and Economic Implications. 16 Dec. 2021, 10.1101/2021.12.14.472674. Accessed 8 Mar. 2022.