Organ-on-a-Chip technology continues to mature and gain adoption by the scientific community as a revolutionary tool for creating novel models of human-centric biology. In this ChipChat blog, we spoke with Lorna Ewart, Chief Scientific Officer at Emulate, to better understand the many exciting—and unexpected—ways Organ-Chips are being used and why her favorite Organ-Chip model may or may not be the Big Toe-on-a-Chip!
Hi Lorna, thank you for taking the time to sit down with us today and discuss the current state of Organ-on-a-Chip technology! I’d like to start by asking you about your “favorite” Organ-Chip model. Whenever someone asks you about the limits of Organ-on-a-Chip technology, you always respond with this idea about creating a “Big Toe-on-a-Chip.” Can you explain why you like to use it as an example?
Lorna Ewart: I use the Big Toe-on-a-Chip for two reasons: Firstly, the Human Emulation System is truly an open-source platform where the only real limit is that of our imagination. We see this across our customer base, where they have developed over 30 different models or applications that have in turn led to over 100 peer-reviewed publications.
The second reason is a bit closer to home. I’ve now been in the field for over 12 years. When I first started to describe what I was doing and how one might approach a “new model,” I found that, whichever organ I referred to, someone would raise a hand and say it had been done. So, when I am now trying to inspire or talk about something without being drawn into organ biology specifics, I refer to “Big Toe-on-a-Chip”. One of these days, someone is going to present me with one, unless of course I get there first!
It’s funny to talk about Big Toe-on-a-Chip, but at the same time, it’s also pretty awe-inspiring to think about how far in vitro modeling technology has come in recent years. Can you explain how Organ-on-a-Chip technology compares to other in vitro models? What makes it better, and why is it considered an advancement in in vitro modeling?
LE: It is generally accepted that scientists first started culturing cells outside of the human body at the turn of the 20th century. At this time, scientists were inspired to explore how a cell may function but had little true knowledge about how to maintain them outside of the body. Consequently, we eventually saw immortalized cell lines being placed on plastic. Given that these cells were typically of a cancerous origin, where they proliferate readily—even on plastic, scientists didn’t need to think about creating an ideal home-away-from home that accurately represented the in vivo environment. I don’t know about you, but there is not a single cell in my body that grows on plastic!
As time progressed, and primary cells began to be isolated from human and animal tissues, scientists recognized that to grow and survive, the plastic had to be coated with a protein to resemble the extracellular matrix (ECM). Thus, primary cell culture was born. The next major discovery was that cellular shape was an important cue, which heralded in the era of spheroids and, ultimately, organoids.
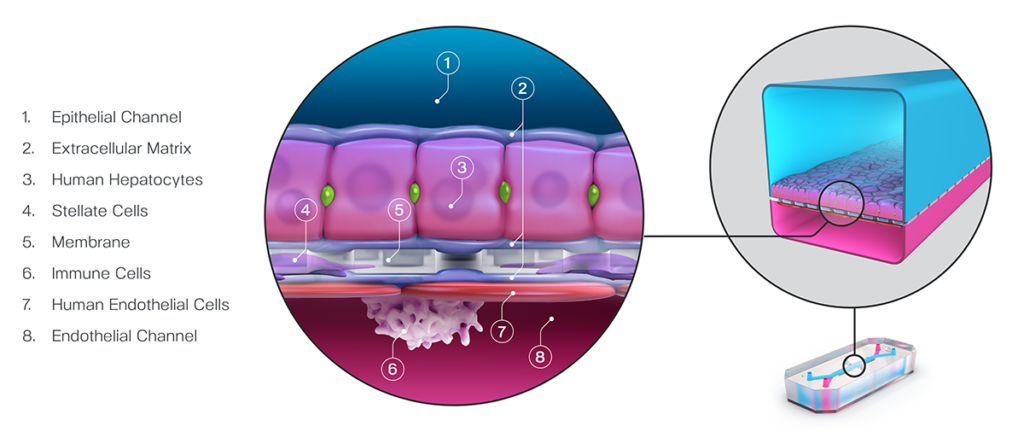
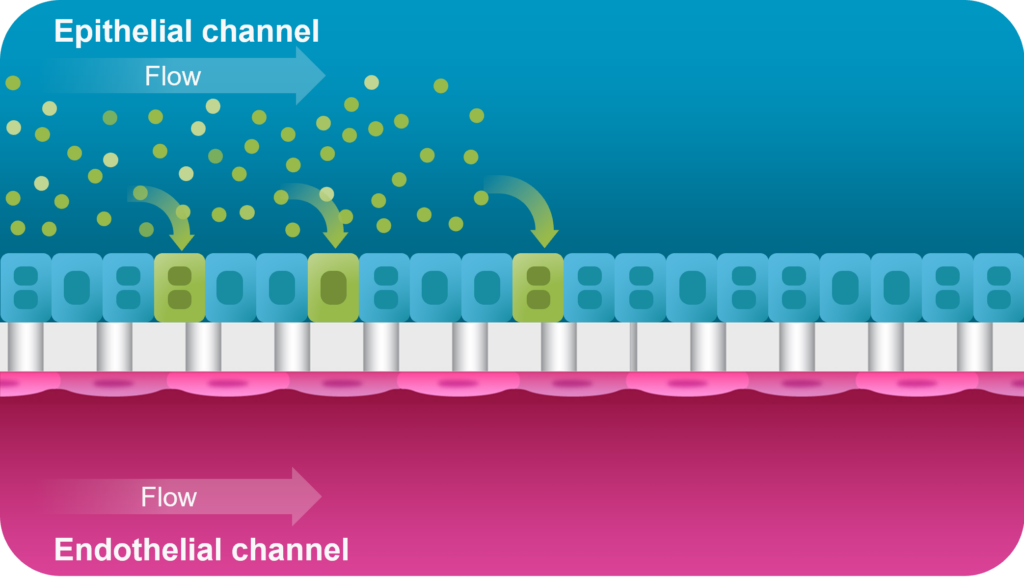
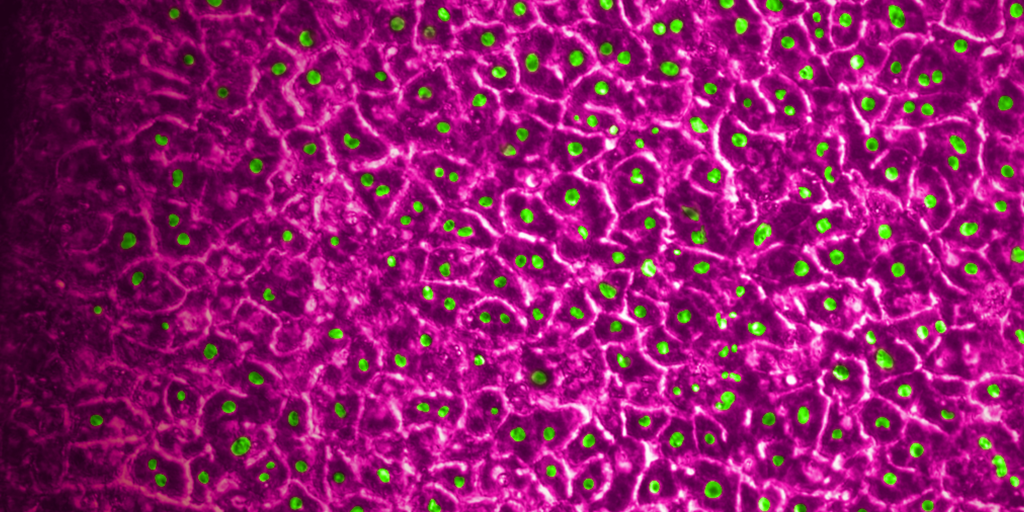
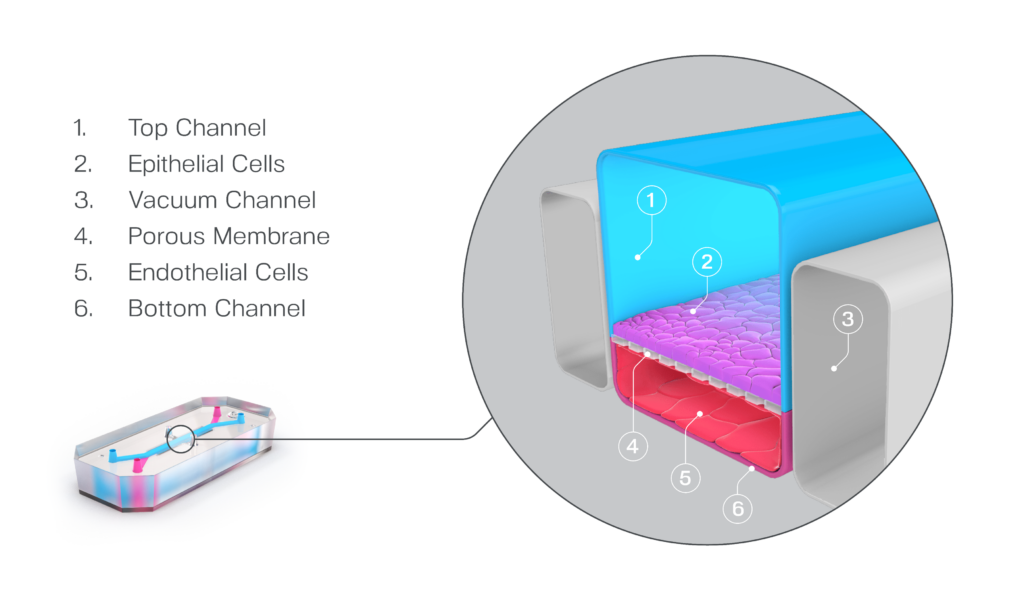
What sets Organ-on-a-Chip technology apart from these other models is that, in addition to ECM and shape, their cells are continually perfused with cell culture media, which replenishes nutrients, allows for waste removal, and enables flow rates high enough to induce shear stress (an important physiological stimulus). Furthermore, Emulate’s microfluidic platform, Zoë Culture Module, can apply vacuum stretch to emulate additional mechanical cues when necessary, such as alveolar expansion and recoil as well as intestinal peristalsis.
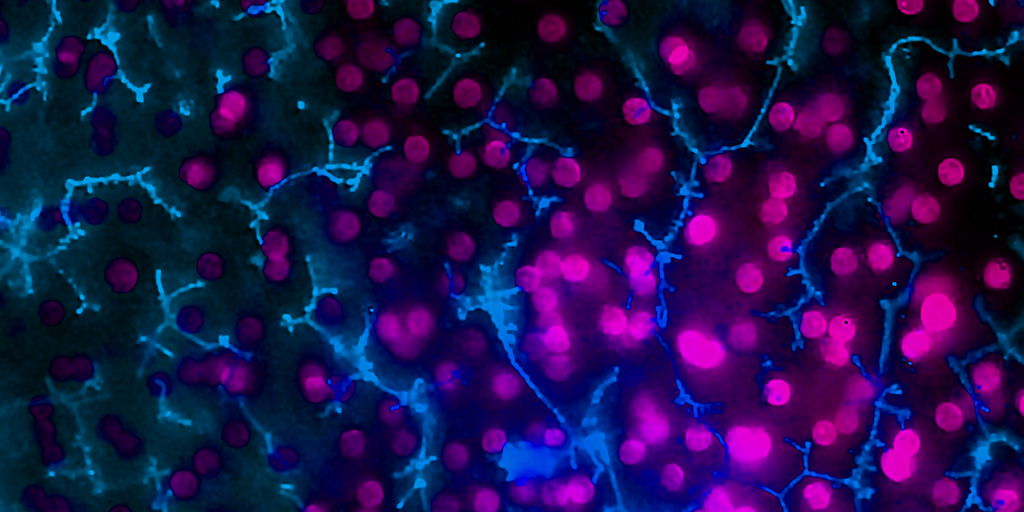
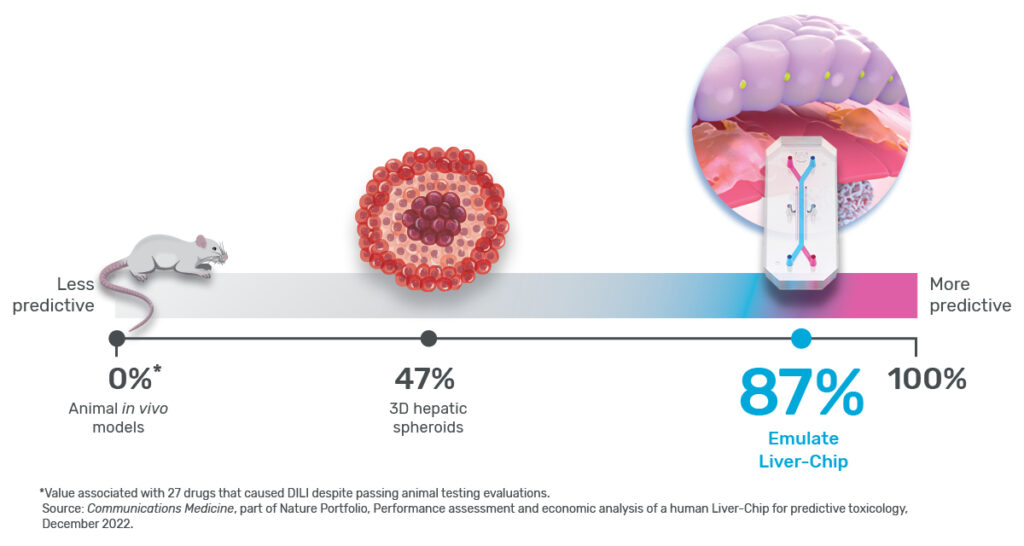
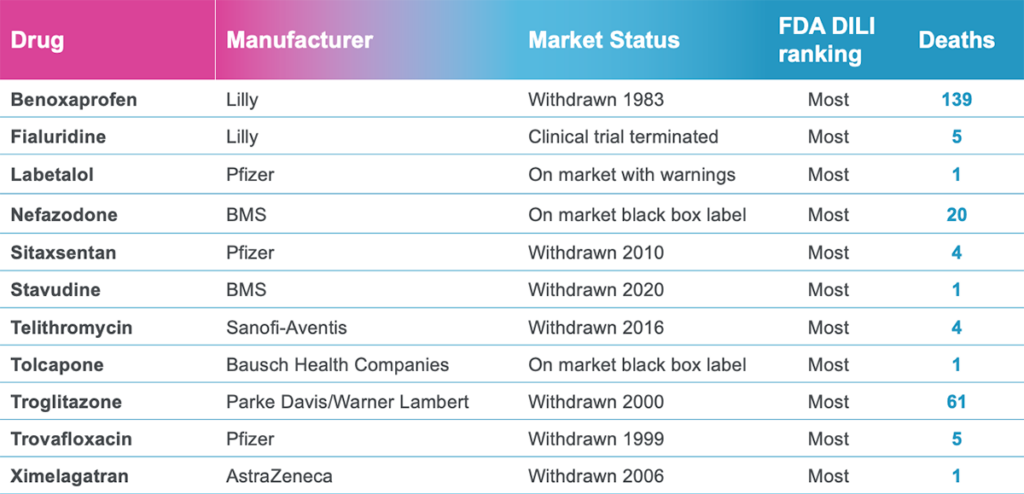
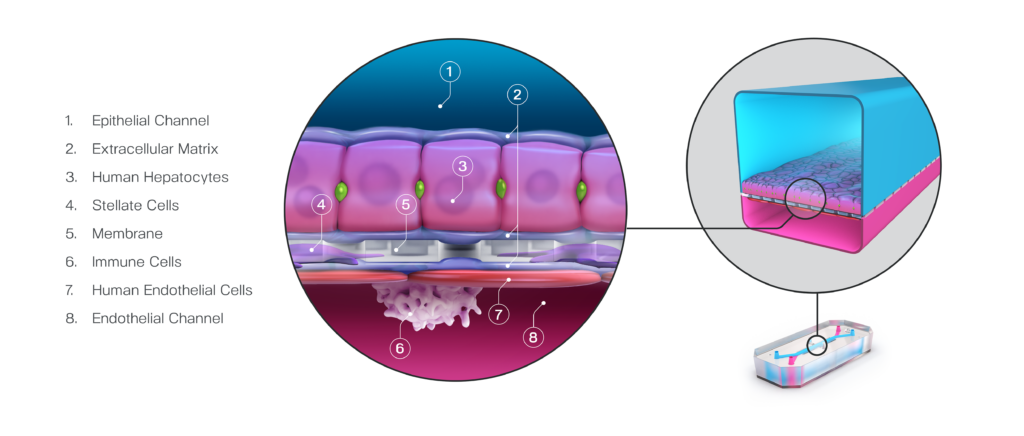
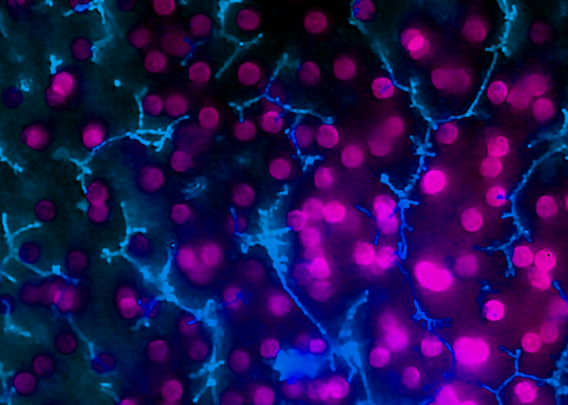
Taken together, these capabilities enable Emulate’s platform to achieve an unparalleled level of in vivo relevance compared to other in vitro models. Of course, the closer a scientist can get to emulating the in vivo microenvironment, the more likely it is that their cells will respond and function in a physiologically relevant manner. In turn, their data sets will have a higher translational value. For example, our landmark findings published in Communications Medicine showed that the Liver-Chip S1 outperformed both animal and spheroid models in predicting drug-induced liver injury for a subset of drugs that failed clinical trials or experienced on-market failures due to toxicity.
What are some of the most unique Organ-Chip ideas you’ve heard? Have you been surprised by any of the asks we’ve received?
LE: With our broad instrument deployment across multiple laboratories in all corners of the world, Emulate is continually seeing a large of number of novel Organ-Chip models being created or adapted for applications like infectious disease and cancer progression. As a female, I am thrilled to see development of Organ-Chip models of the female reproductive system such as the Vagina-Chip and Cervix-Chip, which I hope will in turn lead to new, efficacious treatments for women—something that continues to sadly be overlooked.
On the topic of the Vagina-Chip—I remember it received a lot of attention when that publication came out at the end of 2022. The New York Times even wrote an article about it! Why do you think there was so much buzz around it, and is this an example of how Organ-on-a-Chip technology can help transform the industry’s approach to understanding human health and disease?
LE: Firstly, I believe the buzz was in part because we were normalizing the use of words associated with the female reproductive system, which demonstrates a maturing of scientific thinking beyond developing cancer cures for mice or therapeutics for a 70-kg male. But, moreover, I think the buzz has helped to draw attention to female-specific diseases that have been woefully underfunded for decades. For example, bacterial vaginosis affects up to 25% of reproductive-age women. These bacteria are often resistant to antibiotics, enabling them to thrive, resulting in inflammatory episodes that can sadly lead to pelvic and endometrial diseases. It is amazing to see that, in Prof. Don Ingber’s lab, the co-development of the Vagina- and Cervix-Chips are enabling foundations such as the Bill and Melinda Gates foundation to treat women in Africa, where bacterial vaginosis is highly prevalent and can lead to increased HIV infection while also increasing the chance of pre-term labor.
You’ve been with Emulate for over four years now. What was your vision for the technology when you first started, and where do you see the technology heading now?
LE: When I joined Emulate, I envisioned a future where every lab across the world would have a Zoë. Although we’ve not quite got global domination (yet), we do have hundreds of Zoës in the field working hard to deliver new biology and develop exciting new therapeutics. However, what I find fascinating is the evolution of the consumables that work with Zoë.
When I started, we had our flagship Chip-S1® Stretchable Chip, which has the two parallel microchannels housed in a PDMS body. Through the dedication and talent of our engineering team, we now have the Chip-R1™ Rigid Chip, which has all the hallmarks of Chip-S1 (aside from stretch) but with minimal drug absorption risk and the potential for a higher, physiologically relevant vascular shear stress. I can’t wait to see how this will benefit our customers in ADME research, where the risk of drug absorption can hamper data interpretation. I’m also excited to see this consumable’s potential in the vascular biology community, where we can extend the promising immune cell recruitment application, which includes modeling the functional spectrum of CAR T therapies (e.g., attachment, migration, and killing).
And we must not forget the third chip in our family, the Chip-A1™ Accessible Chip. Inspired by the Transwell, this chip contains an accessible central chamber capable of housing a 3 mm gel, enabling biologists to extend their modeling capabilities of complex tissues such as the tumor microenvironment (TME). Moreover, the accessibility of this chip will enable substances to be directly applied to cells, such as in aerosolization or dermal applications.
You mentioned that we haven’t yet achieved world domination. It’s exciting to see the amount of progress Organ-Chips are making in the field, but what do you think is holding people back from adopting the technology? What can we do to help scientists overcome that “activation energy” hurdle, so to speak?
LE: Yes, it is easy to get carried away with the possibilities, but you are correct, it is also important to focus on overcoming the “activation energy” hurdle. To do this will take a big collaborative effort across the wider community, but there are three things which I believe are essential and will move the needle.
The first is for us and other Organ-Chip developers to show consistent innovation in our platform and consumable offerings. I’ve already mentioned the consumables, but it is important to understand how to reduce the instrument complexity while leaving the only complexity to the biological model. The second aspect would be to generate a series of large data sets, not unlike our aforementioned work with the Liver-Chip. Such data sets will enable true characterization of the system and biology, ultimately showing the possibilities and likely limitations which can be cycled back into R&D teams.
Lastly, I believe the regulatory community plays a key role in promoting adoption. The US Food & Drug Administration (FDA) launched its Innovative Science and Technology Approaches for New Drugs (ISTAND) Pilot Program to help facilitate the qualification of drug discovery tools, of which Organ-Chips are a great example. In September, we were able to share that Emulate’s Liver-Chip S1 had been accepted into the ISTAND program, the first Organ-Chip to achieve this honor. We are working hard on our qualification plan and are excited to be collaborating with the FDA on this momentous journey.
It sounds like a promising step towards the vision of having Organ-Chips in every lab! On a final note, let’s say that someone reading this blog does want to model a Big Toe-on-a-Chip. In all seriousness, are there any practical applications of creating a Big Toe-on-a-Chip? And if someone wanted to model a Big Toe–on–a–Chip, how would they go about that?
LE: So, aside from causing ridiculous pain when you stub it on the corner of the bed, the big toe actually does perform a very important role! It is four times bigger than other toes and contains more muscles, meaning it is vital for balance. Lovers of fashion shoes, especially of the high-heeled variety, can develop bunions, which are incredibly painful. So, maybe, the Big Toe-on-a-Chip can shed light on what “changes” in this vital body part to lead to a condition that is generally addressed with surgery. Separately to this, gout and arthritis commonly affect the big toe. Both conditions are also incredibly painful, so using this type of model to discover new, potentially locally active treatments would bring relief to many. Because my biggest passion is ultimately ensuring a high quality of life for humankind, perhaps I need to get my thinking cap on to make this model a reality.