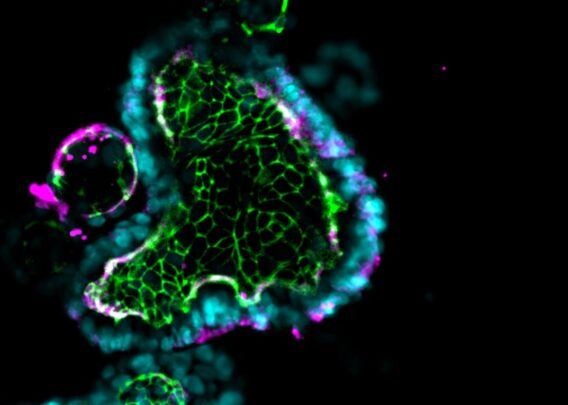
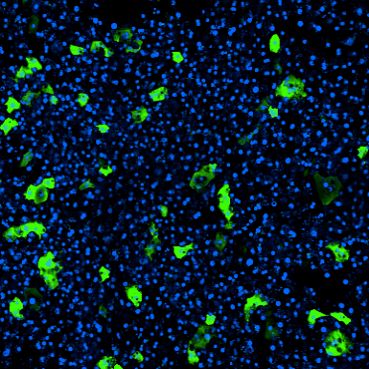
In our new ChipChat™ Q&A Series, we’ll be taking you into the lab with one of our expert scientists to see exactly how Organ-Chips are used and why they improve research. In this interview, Emulate Director of Immunology Chris Carman, PhD, discusses our new immune cell recruitment application for the Colon Intestine-Chip and how it can enable researchers to study complex immune response with unprecedented physiological relevance.
What is immune cell recruitment?
Chris Carman: Our immune system monitors for signs of danger, damage, and infection in the human body. Immune cells travel through the blood stream until they are recruited to enter infected tissue, after which they will respond to the infection, clear it, heal the tissue, and then either die or exit back to the bloodstream. As you can imagine, it is very important that this process be tightly regulated and self-limited; if it weren’t, we’d suffer from all kinds of autoimmune diseases starting at a very early age. So, for the immune system to function properly and to maintain health and homeostasis, the selectivity of immune cell recruitment and response is essential.
Why is immune cell recruitment important when researching inflammatory bowel disease (IBD) as well as other diseases?
CC: For the vast majority of us, a vast majority of the time, the immune system functions precisely as I just described: recruited immune cells travel to a particular location with a specific purpose. When their job is complete, these immune cells clear out, and the inflammation subsides.
For those afflicted with inflammatory bowel disease (IBD), however, the process I just described becomes dysregulated, and immune cells are excessively recruited to unintended locations. This causes further dysregulated immune cell recruitment and reactions, triggering a vicious cycle of pro-inflammatory responses that ultimately cause tissue damage and dysfunction, leading to disease.
The truth is that dysregulated, excessive inflammation is at the heart of all major human diseases. IBD is an excellent example of a disease that depends on, and is ultimately driven by, excessive, dysregulated immune reactions.
What is Emulate currently doing with immune cell recruitment, and how does that speak to the process you just discussed?
CC: IBD is a complex, chronic disease that is incredibly difficult to study. Its hallmark, the disruption of the epithelial barrier, causes materials inside the intestine, including bacteria, to leak. You can imagine how that drives a subsequent escalation of inflammation as the cycle gets kicked off. What we at Emulate have done in a, frankly, unprecedented manner is model the entire process. As inflammation starts, there is always a priming cue that causes local tissue to undergo changes, including critical pro-inflammatory reprogramming of endothelial cells lining the vessels. Our model begins precisely at this priming step, and it then goes on to capture the full course of the disease progression.
“The truth is that dysregulated, excessive inflammation is at the heart of all major human diseases. IBD is an excellent example of a disease that depends on, and is ultimately driven by, excessive, dysregulated immune reactions.”
chris carman, phd
Could you go into a bit more detail about your new immune cell recruitment application and how it improves upon previous models?
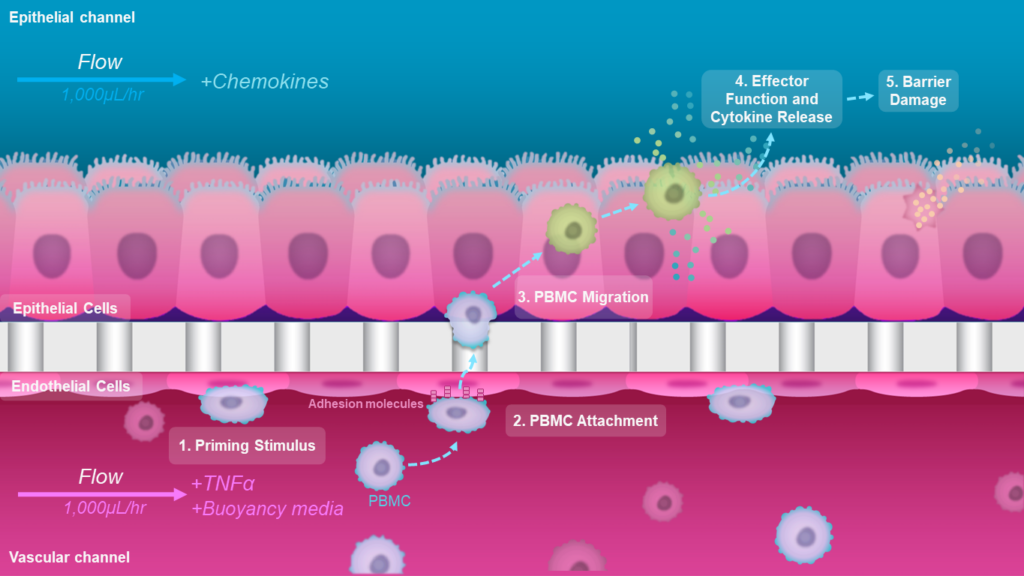
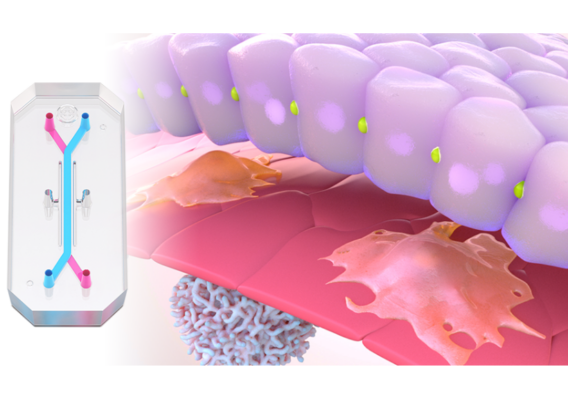
CC: To develop this application, we used the Colon Intestine-Chip, which is a primary human cell model of the colonic barrier that co-cultures organoid-derived epithelium with colon-specific vasculature. We demonstrate in published work that the morphology, function, and transcriptome signature of this model very tightly recapitulate human physiology in a manner that is dependent upon the cell-cell interactions with vascular endothelial cells. This feature is unique to this model compared to competing technologies that lack vasculature, such as organoid-based approaches.
To initiate inflammation, we applied a well-established cytokine that drives the early IBD progression as the priming stimulus. Critically, we next introduced immune cells—specifically, peripheral blood mononuclear cells (PBMCs)—into the Colon Intestine-Chip’s vascular channel. With this step, our model could capture the complexity of human pathophysiology and was able to recapitulate most of the critical sequence of events for IBD, including immune cell adhesion to the endothelium and migration into the tissue, activation of complex interstitial immune signaling networks, and finally a release of the critical hallmark cytokines and disruption of the epithelial barrier.
How will this model benefit IBD research?
CC: First, a critical aspect of this model is that it represents a more complete and complex recapitulation of both human intestinal physiology and disease. We believe that this unprecedented completeness coupled with experimental tractability will lead to a deeper mechanistic understanding of IBD. Second, and equally as important, we believe this model will enable researchers to identify new therapeutic IBD targets more precisely so that better and more effective therapeutics can be developed and validated. Ultimately, we hope that this model will help researchers greatly diminish the attrition rate of drugs moving into the clinic.
How does the Emulate immune cell recruitment application improve upon traditional models of IBD?
CC: Traditional models used to study IBD and develop therapeutics for the disease have yielded significant learnings, but they also exhibit important limitations. For example, conventional in vivo studies are almost always performed in mice, which suffer from species-specific differences—a factor that is particularly important for studying the immune system. At the same time, traditional in vitro models involving epithelial cell lines or organoid cultures are highly constrained by their limited complexity. Each of these models has strengths and weaknesses, but they are really only able to look at one piece of the puzzle, and none captures the full complexity of human disease. As such, researchers can only capture a subset of therapeutic targets with these models. Because the Colon Intestine-Chip and immune cell recruitment application capture a more complete sequence of events for IBD, researchers can study a much broader spectrum of disease targets and, subsequently, develop more effective therapeutics.
What other ways can Organ-Chips be used for immunology beyond this application?
CC: In addition to modeling circulating immune cell recruitment, researchers can incorporate so-called resident immune cells—which also play essential roles in driving immune response—into Organ-Chip models. A couple of our developed models apply this functionality today: The Liver-Chip incorporates Kupffer cells to enable studies of immune-mediated toxicity of drug candidates; and the Brain-Chip incorporates microglia to enable studies of neuroinflammation—a process that is implicated in many neurodegenerative diseases, including Alzheimer’s and Parkinson’s disease.
Researchers at the Wyss Institute have also modeled the immune system by creating a Lymphoid Follicle-Chip, which they have used to recapitulate human immune function and evaluate the efficacy of vaccines for the flu and COVID-19.
How can researchers gain access to these new capabilities and use them in their own work?
CC: Researchers can gain access to these new capabilities in a couple of ways. First, they can work with our in-house service team of experts who can design and execute a study to investigate the efficacy or toxicity of anti-inflammatory drug candidates for IBD. Second, they can bring Emulate Organ-on-Chip technology—which we call the Human Emulation System®—into their own labs. We offer instrumentation to automate cell culture conditions, Bio-Kits that include the chips and primary human cells customers need to build the Colon Intestine-Chip, and robust protocols, training, and experimental support to help drive success. Whether the research is performed in our labs or in our customer’s labs, our hope is that these new capabilities can enable researchers to develop novel, more effective therapeutics for IBD.