Using a Human-Relevant Model
Shigella are bacteria that cause Shigellosis, a gastrointestinal disease inducing severe stomach cramps, fever, and diarrhea. The onset of symptoms can begin from 1-2 days of contact and results from the rapid invasion of the pathogen within intestinal epithelial cells, leading to the destruction of healthy human colon tissue. In developing countries, Shigellosis remains an important health threat and can lead to epidemics where sanitation is poor[1]. Every year, Shigellosis kills hundreds of thousands of individuals, mostly children younger than the age of five[2].
It is challenging to study this pathogen because Shigella infects only humans, resulting in colon infection. Therefore, small animal models, such as mice, cannot be used to study the disease. Also, conventional cellular in vitro models, such as human polarized epithelial cells grown on transwells, fail to develop efficient infection. Thus, it has been difficult to study the inception of Shigella infection and understand how this pathogen interacts with the epithelial barrier at the tissue scale. For decades, the scientific community has been missing a reliable, consistent, and clinically relevant Shigella infection in a polarized intestinal epithelial monolayer.
Leveraging Organs-on-Chips technology from Emulate, researchers published a paper in Cell Host & Microbe, highlighting how a human Intestine-Chip can be used to test the infectivity of Shigella. The paper showcases a number of experimental approaches such as real-time imaging, tissue cross-section analysis, and conventional microbiology.
Creating the Intestine-Chip for Shigella Testing
First, the microenvironment of the human intestine was recreated by utilizing Organs-on-Chips technology to create an Intestine-Chip model. The chip itself is made of an elastomeric polymer which consists of a central channel separated horizontally by a flexible porous membrane; two hollow vacuum channels laterally frame the central channel. The application of cyclic vacuum within these lateral channels induces strain and stretching of the porous membrane that recreate peristaltic-like motions (in vivo-relevant mechanical forces). In order to support the culture of human colonic epithelial cells, Caco-2 cells were used, and the separating membrane was coated with extracellular matrix (ECM). To recreate the physiological microenvironment of the intestine, the seeded cells were exposed to continuous flow for 5-6 days to recreate the shear stress on the epithelial channel and cyclic mechanical strain to reproduce the peristaltic motions.
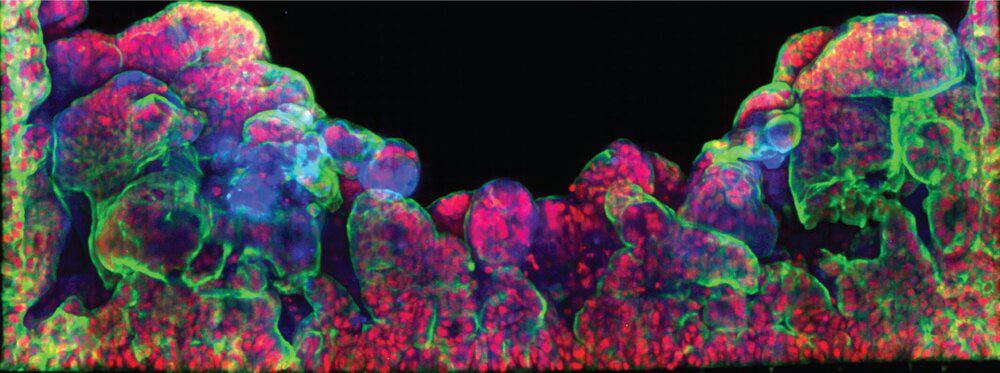
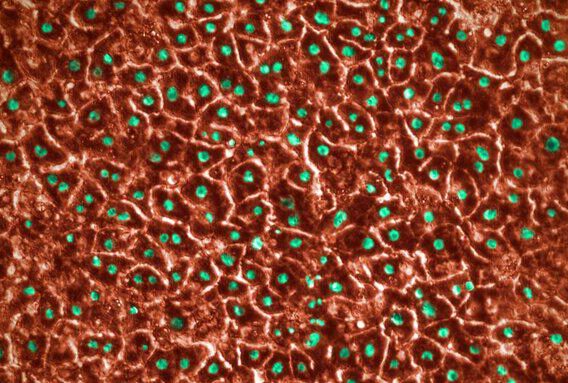
Cells grown under these conditions self-organized into villi-like structures interspaced by crypt-like invaginations. Confirming that the seeded Caco-2 cells were polarized and exhibited known differentiation markers (E-cadherin, villin, and FABP1-KRT20). Scanning electron microscopy (SEM) imaging also showed microvilli in the epithelial channel, a typical structural marker of differentiated and polarized enterocytes.
Next, in order to address whether Shigella flexneri could infect the human colon epithelium in the chip, a wild-type strain expressing GFP (Shigella-WT-GFP) in the intestinal lumen of the Intestine-Chip was introduced.
Human-Relevant Data
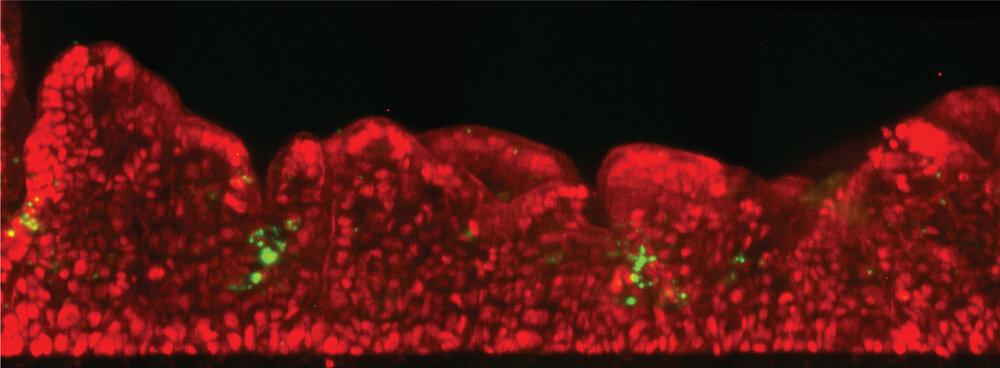
Shigella invasion was observed when only a few hundred bacteria were introduced into the intestinal lumen. The observation that only minimal loads were required for infection is consistent with data from the clinic. Given this correlation between in vivo and chip data, the approach in the chip allowed us to dissect the fundamentals mediating Shigella invasion at the tissue scale. In particular, identification that Shigella directly infect enterocytes from the epithelial channel, shifting the current paradigm about the early stage of invasion. Secondly, the study found that Shigella take advantage of the 3D topology of the tissue to populate specific areas in the colon tissue reminiscent of the human colon crypts. Finally, the design of the Intestine-Chip recreates the mechanical forces on the pathogen invasion and found that peristalsis is critical for specific stages of the infection process.
Most surprising was the observation that Shigella could directly infect enterocytes from the epithelial side (from the intestinal lumen) as expected in vivo with such efficiency and reproducibility.
Collaboration
This work is the result of a collaboration between the Unité de Pathogénie Microbienne Moléculaire and Biomaterials and Microfluidics Core Facility at Institut Pasteur and Emulate. It exemplifies how joining forces from different areas of expertise (engineering, microbiology, and bio-imaging) can reveal new avenues to better understand the mechanism of infection by enteroinvasive bacteria. It also helps advance our understanding of human disease, ultimately enabling the development of new therapeutics.