Organ Model: Esophagus
Application: Cancer
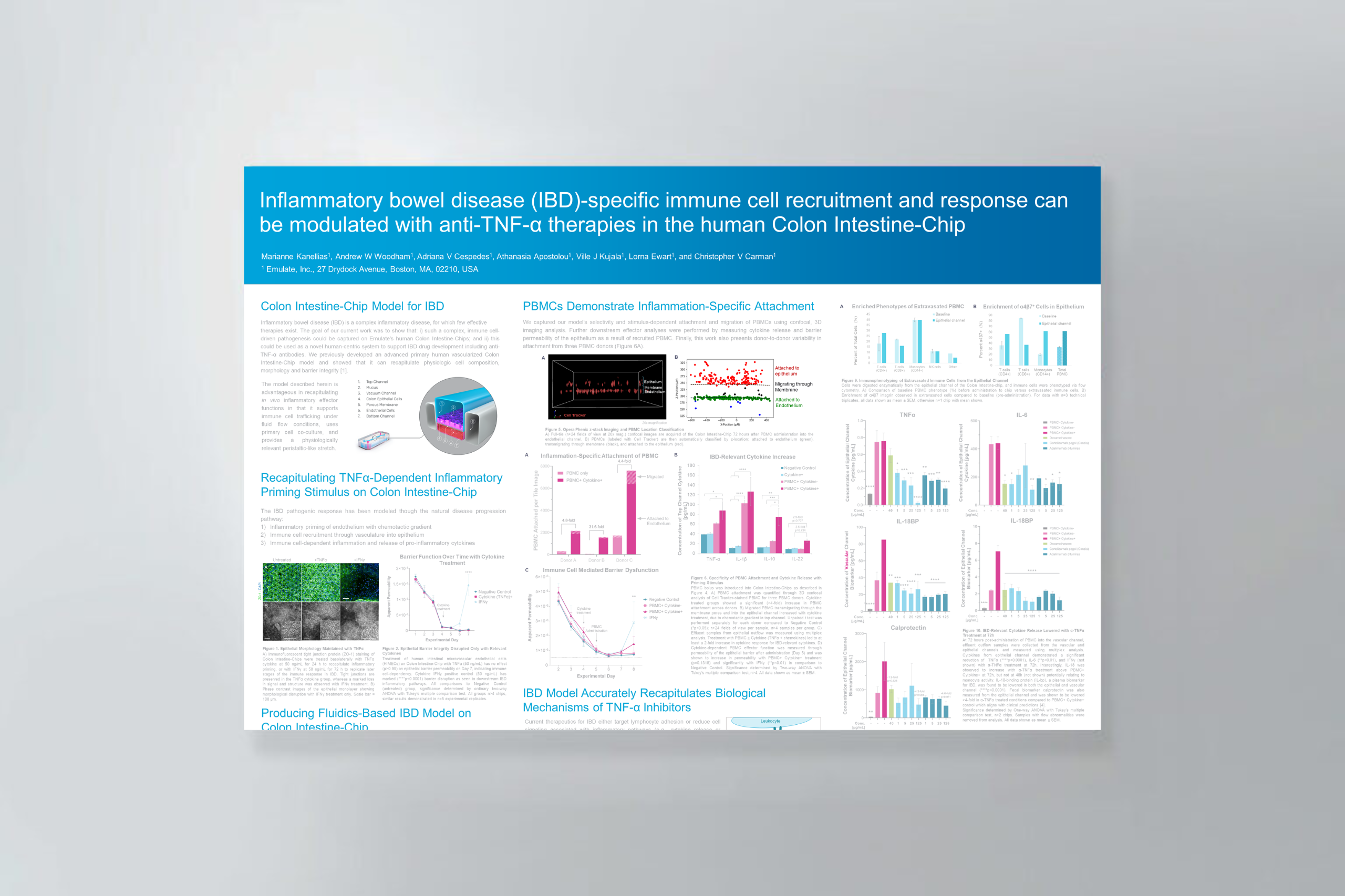
Abstract: The pathogenesis of subsquamous intestinal metaplasia (SSIM), in which glands of Barrett’s esophagus (BE) are buried under esophageal squamous epithelium, is unknown. In a rat model of reflux esophagitis, we found that columnar-lined esophagus developed via a wound-healing process involving epithelial-mesenchymal plasticity (EMP) that buried glands under ulcerated squamous epithelium. To explore a role for reflux-induced EMP in BE, we established and characterized human Barrett’s organoids and sought evidence of EMP after treatment with acidic bile salts (AB). We optimized media to grow human BE organoids from immortalized human Barrett’s cells and from BE biopsies from seven patients, and we characterized histological, morphological, and molecular features of organoid development. Features and markers of EMP were explored following organoid exposure to AB, with and without a collagen I (COL1) matrix to simulate a wound-healing environment. All media successfully initiated organoid growth, but advanced DMEM/F12 (aDMEM) was best at sustaining organoid viability. Using aDMEM, organoids comprising nongoblet and goblet columnar cells that expressed gastric and intestinal cell markers were generated from BE biopsies of all seven patients. After AB treatment, early-stage Barrett’s organoids exhibited EMP with loss of membranous E-cadherin and increased protrusive cell migration, events significantly enhanced by COL1. Using human BE biopsies, we have established Barrett’s organoids that recapitulate key histological and molecular features of BE to serve as high-fidelity BE models. Our findings suggest that reflux can induce EMP in human BE, potentially enabling Barrett’s cells to migrate under adjacent squamous epithelium to form SSIM.NEW & NOTEWORTHY Using Barrett’s esophagus (BE) biopsies, we established organoids recapitulating key BE features. During early stages of organoid development, a GERD-like wound environment-induced features of epithelial-mesenchymal plasticity (EMP) in Barrett’s progenitor cells, suggesting that reflux-induced EMP can enable Barrett’s cells to migrate underneath squamous epithelium to form subsquamous intestinal metaplasia, a condition that may underlie Barrett’s cancers that escape detection by endoscopic surveillance, and recurrences of Barrett’s metaplasia following endoscopic eradication therapy.
To learn more about these findings, view our webinar “Organ-Chips 201: The Importance of Flow, Stretch, and Stroma for in vitro Modeling.”