Version 1.0 (Release Date: March 2022)
Introduction
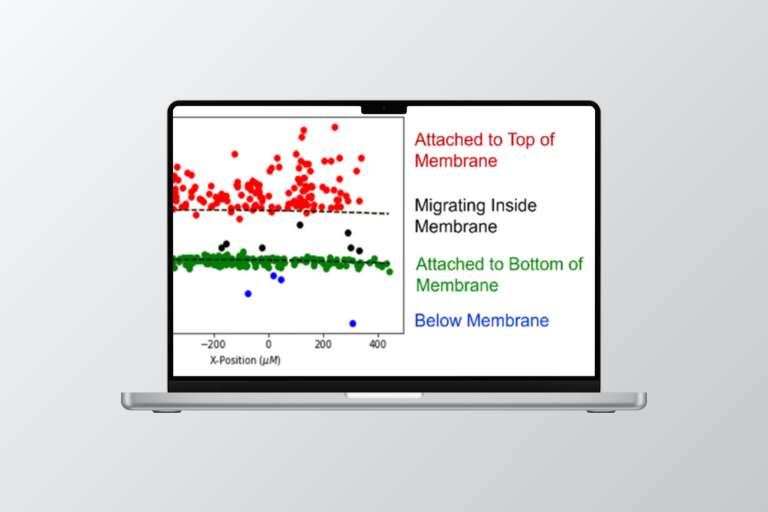
These Compound Distribution Kit analysis tools simplify assay preparation and data analysis for the Emulate Compound Distribution Kit (CDK) protocol, enabling users to quantify the amount of test compound loss (e.g., absorption or adsorption) ahead of a Chip-S1® Stretchable Chip experiment.
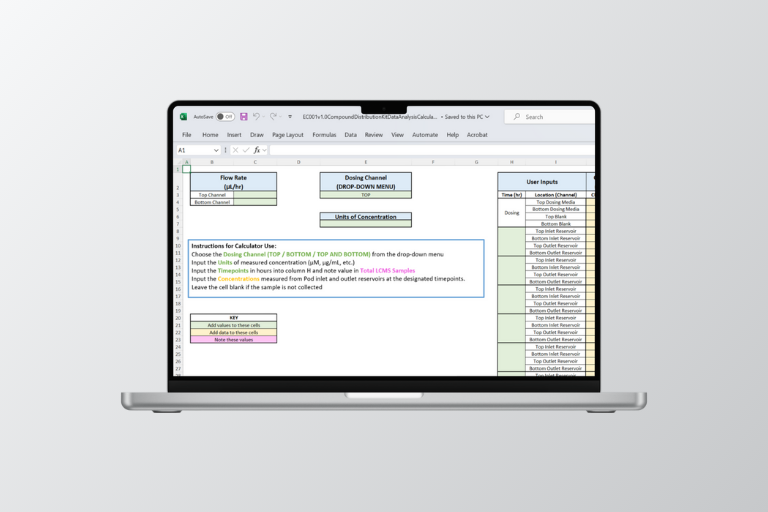
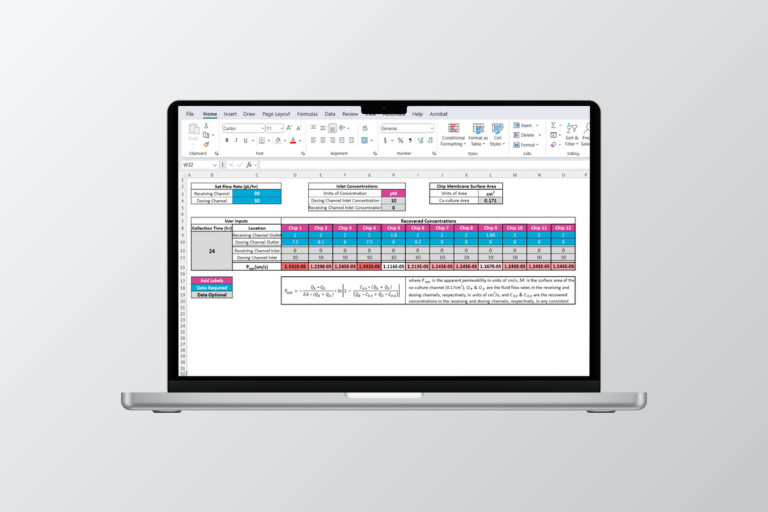
There are two Excel-based tools in the download: The Media Volume Preparation Calculator and the CDK Data Analysis Calculator. The Media Volume Preparation Calculator calculates the media volume required for the CDK assay, based on the intended flow rate, study duration, and expected re-dosing protocol. The CDK Data Analysis Calculator calculates the adjusted average cellular exposure and concentration range based on the dosing concentration.
Related Information: