Organ Model: Intestine
Applications: ADME
Highlights
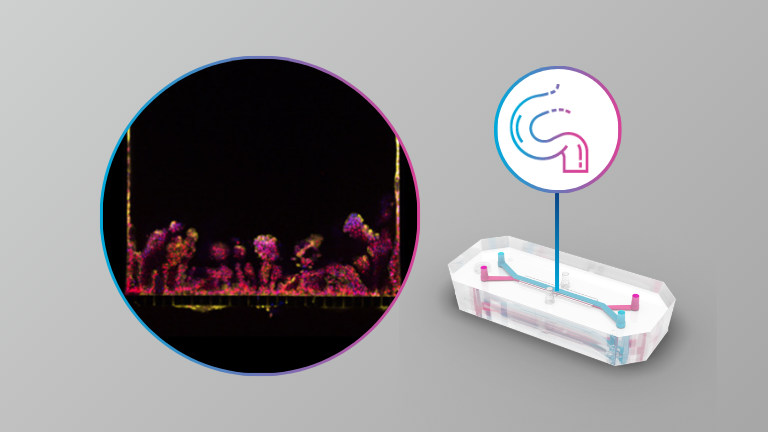
This study directly compared three culture formats—organoids, Transwells, and intestine-on-chip systems—using the same hiPSC-derived intestinal epithelial cells to understand how culture context shapes biology. While all three models supported intestinal differentiation, the Intestine-Chip consistently reproduced the most mature, adult-like phenotype, with the strongest upregulation of genes involved in digestion, nutrient transport, and drug metabolism, including CYP enzymes. The dynamic flow and physiologically relevant microenvironment provided by the Organ-Chip drove functional maturation that static organoid and Transwell systems could not achieve. These findings highlight Organ-Chips as a more predictive platform for studying human intestinal function, drug absorption, and DDI risk.